Single cell expression analysis uncouples transdifferentiation and cell reprogramming
Two types of cell plasticity
Some somatic cell types are plastic, having the capacity to convert into other specialized cells (transdifferentiation) or into induced pluripotent stem cells (iPSCs, reprogramming) in response to transcription factor over-expression. However, cellular transdifferentiation and reprogramming efficiencies are highly variable across different cell types.
Single cell analyses of induced B cell conversions reveals asynchronies
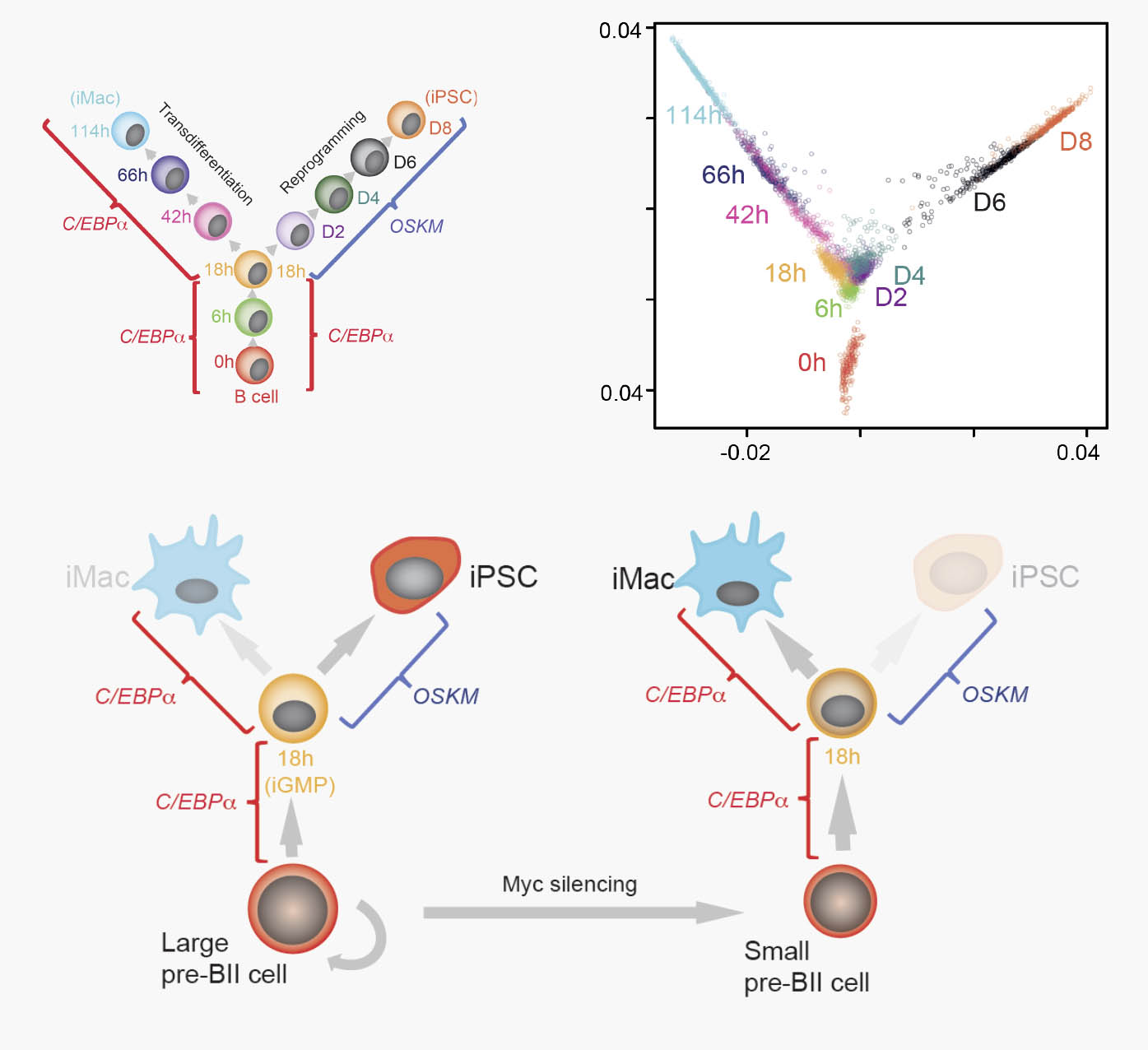
Some somatic cell types are plastic, having the capacity to convert into other specialized cells or into iPS cells in response to transcription factor over-expression. However, cellular transdifferentiation and reprogramming efficiencies are highly variable across different cell types. To determine whether cells highly responsive to induced transdifferentiation are also easily reprogrammable, we exposed bone marrow derived pre-B cells to two different transcription factor overexpression protocols that convert them either into macrophages or iPSCs and monitored the two processes over time using single cell gene expression analysis. We found that although cell conversion trajectories were largely homogenous, cells at some intermediate stages varied in both their speed and path of transdifferentiation and reprogramming.
Large and small pre-B cells differ in their susceptibilities to transdifferentiation and reprogramming, correlating with their initial Myc levels
Unexpectedly, this heterogeneity originates in the starting cell population, which consists of a mixture of large pre-BII cells and small pre-BII cells. The large cells transdifferentiate slowly but exhibit a high efficiency of iPSC reprogramming. In contrast, the small cells transdifferentiate rapidly but are resistant to reprogramming. The large cells exhibit high Myc activity and are cycling whereas the small cells are Myc low and mostly quiescent. Our data reveal that the rate and path of cell fate conversion vary even in highly efficient systems and that this relates to variation in the starting cell state. They also show that a somatic cell’s propensity for transdifferentiation and reprogramming are uncoupled and illustrate how single cell expression and computational analyses can identify the origins of heterogeneity in cell fate conversion processes. Finally, in published work differences in Myc activity predict the efficiency of iPSC reprogramming across a wide range of somatic cell types.