Role of genome topology during cell reprogramming
Our cells contain the same genetic information, represented by 23 pairs of chromosomes with about 20,000 genes. The several hundred different cell types in our body are specified by the selective expression of genes located in different parts of the genome. The machinery required to selectively recall this information resides primarily in transcription factors, proteins that switch genes on or off. Our recent results now suggest an additional role of transcription factors, namely in shaping the genome, and that genome architecture provides another layer of function involved in cell fate control.
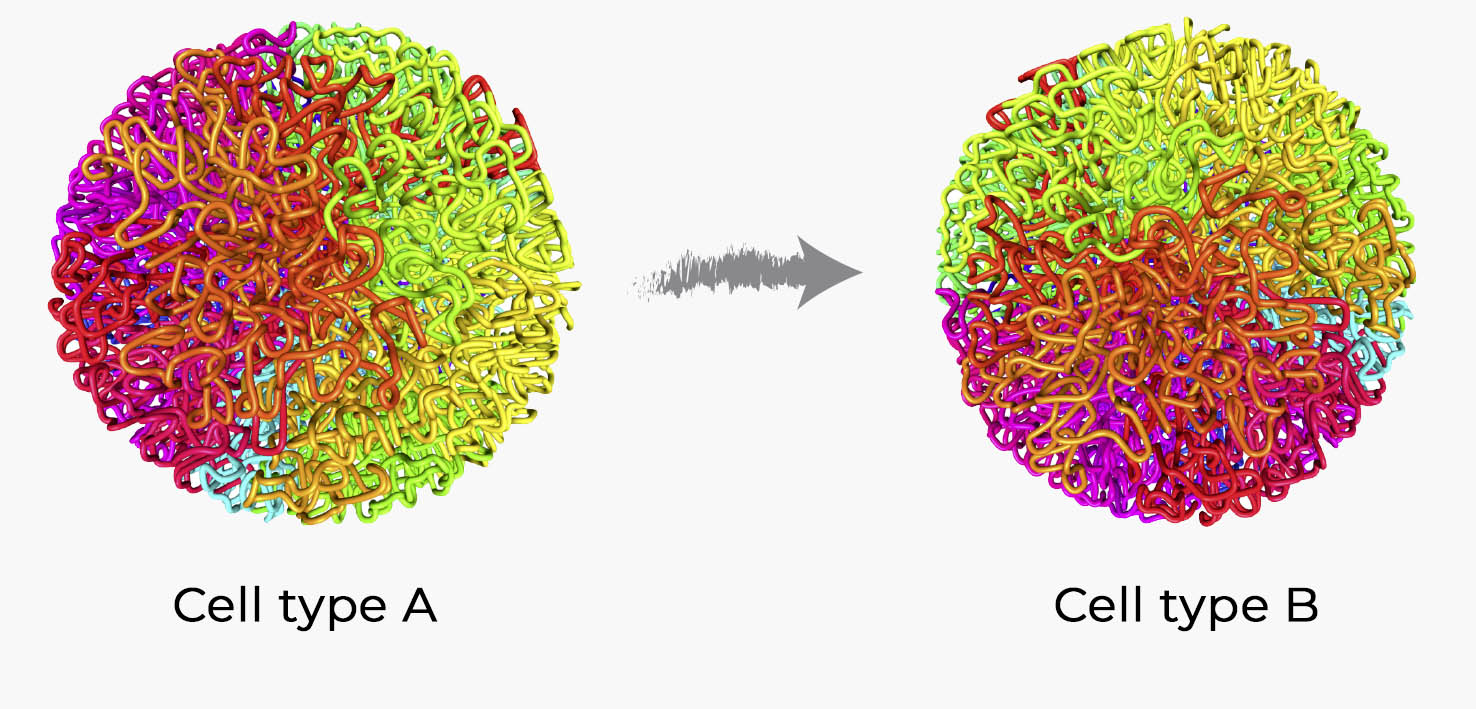
Genome topology changes during reprogramming of B cells into iPScells
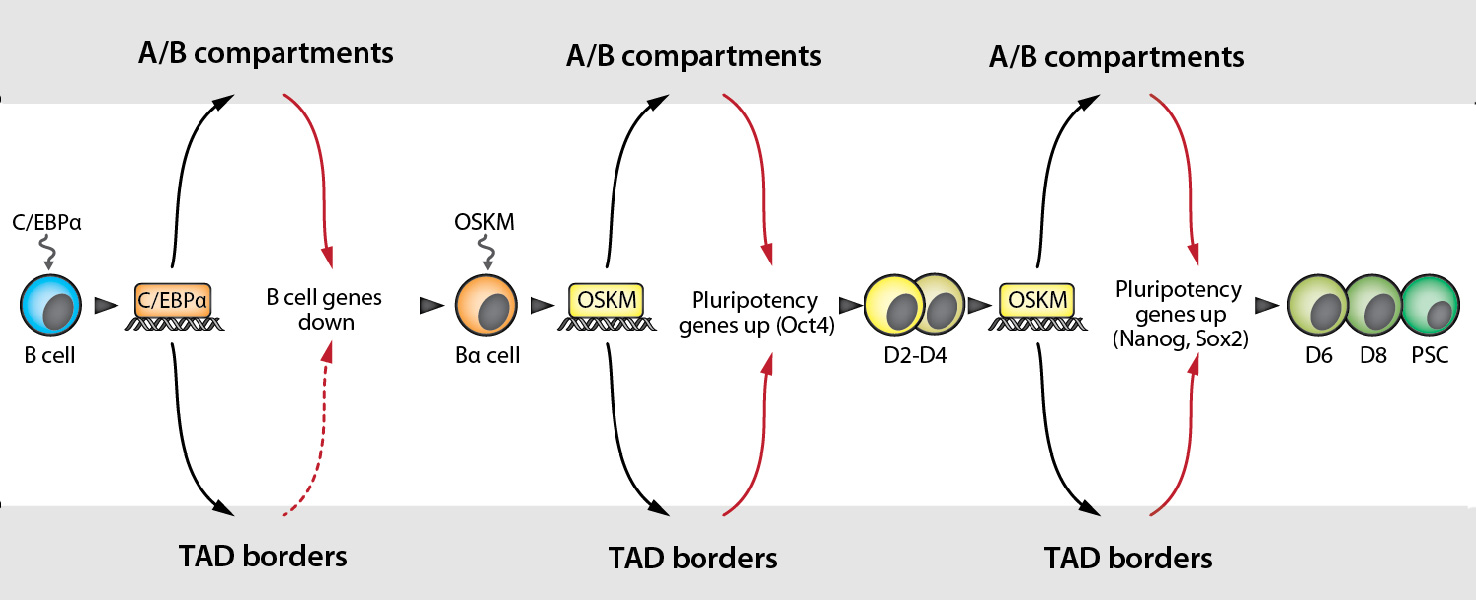
Using a technique called ‘HiC’ that permits mapping contact sites within chromatin revealed that the 3D-architecture of the genome (compartments, TAD borders) is mostly similar between different cells, but that there are also cell type-specific differences. This raises the question: are the architectural differences between cell types merely a consequence of differences in gene expression, or does the 3D organization of the genome itself carry important information for gene regulation? To address this question, we need to observe genome architecture changes during cell fate conversion over time and to compare them to changes in gene expression. To do this we used the rapid and highly efficient reprogramming system developed in the lab, in which pre-B cells are converted into iPS cells by a pulse of C/EBPa followed by the sequential activation of the Yamanaka factors (OSKM).
Transcriptional activation of many genes is preceded by genome topology changes
Surprisingly our findings revealed that, although changes in 3D organization and transcription often occurred at the same time, in many genomic regions architectural changes happened before genes became expressed or silenced, and very rarely the other way around. Within the group of genes that are transcriptionally activated subsequent to changes in genome architecture we detected two (Nanog and Sox2) that encode transcription factors important for the formation of pluripotent stem cells. Our discovery indicates that C/EBPa and the Yamanaka factors directly alter genome architecture and can remove ‘roadblocks’ necessary to create a chromatin substrate permissive for late acting transcription factors. Our findings indicate that genome topology has an informational value for gene expression during cell fate changes and explain the observation that some pluripotency genes only become activated at late stages of reprogramming. These observations are relevant for the formation of pluripotent stem cells during early embryo development as well as for certain developmental malformations of humans due to mutations that cause a derangement of genome architecture.